#include "sat/glucose2/System.h"#include "sat/glucose2/ParseUtils.h"#include "sat/glucose2/Options.h"#include "sat/glucose2/Dimacs.h"#include "sat/glucose2/SimpSolver.h"#include "sat/glucose2/AbcGlucose2.h"#include "base/abc/abc.h"#include "aig/gia/gia.h"#include "sat/cnf/cnf.h"#include "misc/extra/extra.h"
Go to the source code of this file.
Macros | |
| #define | USE_SIMP_SOLVER 1 |
| DECLARATIONS ///. | |
| #define | Gia_CubeForEachVar(pCube, Value, i) |
| #define | Gia_SopForEachCube(pSop, nFanins, pCube) |
| #define Gia_CubeForEachVar | ( | pCube, | |
| Value, | |||
| i ) |
Function*************************************************************
Synopsis [Computing d-literals.]
Description []
SideEffects []
SeeAlso []
Definition at line 1074 of file AbcGlucose2.cpp.
| #define Gia_SopForEachCube | ( | pSop, | |
| nFanins, | |||
| pCube ) |
Definition at line 1076 of file AbcGlucose2.cpp.
| #define USE_SIMP_SOLVER 1 |
DECLARATIONS ///.
Definition at line 42 of file AbcGlucose2.cpp.
Definition at line 1079 of file AbcGlucose2.cpp.

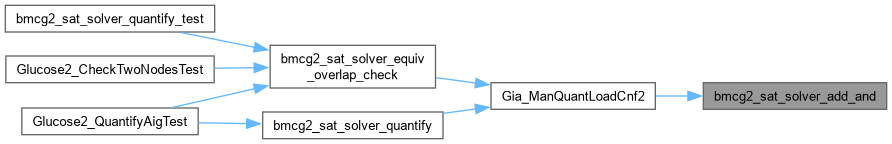
| int bmcg2_sat_solver_add_and | ( | bmcg2_sat_solver * | s, |
| int | iVar, | ||
| int | iVar0, | ||
| int | iVar1, | ||
| int | fCompl0, | ||
| int | fCompl1, | ||
| int | fCompl ) |
Definition at line 316 of file AbcGlucose2.cpp.

| int bmcg2_sat_solver_add_xor | ( | bmcg2_sat_solver * | pSat, |
| int | iVarA, | ||
| int | iVarB, | ||
| int | iVarC, | ||
| int | fCompl ) |
Definition at line 339 of file AbcGlucose2.cpp.

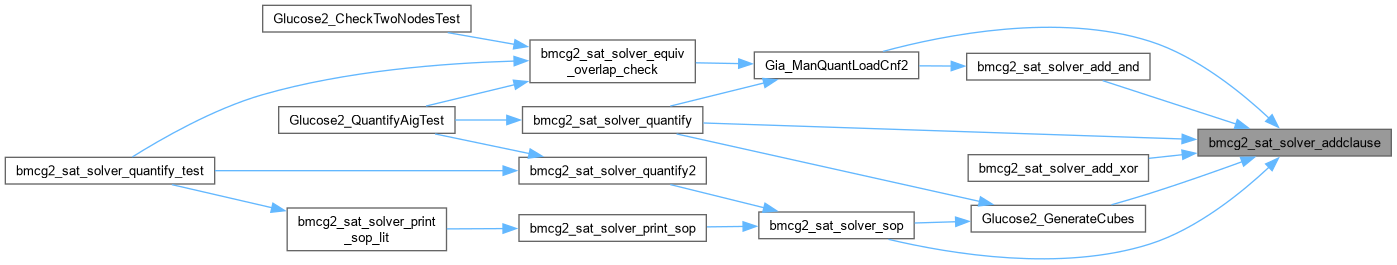
| int bmcg2_sat_solver_addclause | ( | bmcg2_sat_solver * | s, |
| int * | plits, | ||
| int | nlits ) |
Definition at line 165 of file AbcGlucose2.cpp.

| int bmcg2_sat_solver_addvar | ( | bmcg2_sat_solver * | s | ) |
Definition at line 186 of file AbcGlucose2.cpp.

| int bmcg2_sat_solver_clausenum | ( | bmcg2_sat_solver * | s | ) |
Definition at line 259 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_conflictnum | ( | bmcg2_sat_solver * | s | ) |
Definition at line 267 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_elim_varnum | ( | bmcg2_sat_solver * | s | ) |
Definition at line 215 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_eliminate | ( | bmcg2_sat_solver * | s, |
| int | turn_off_elim ) |
Definition at line 198 of file AbcGlucose2.cpp.
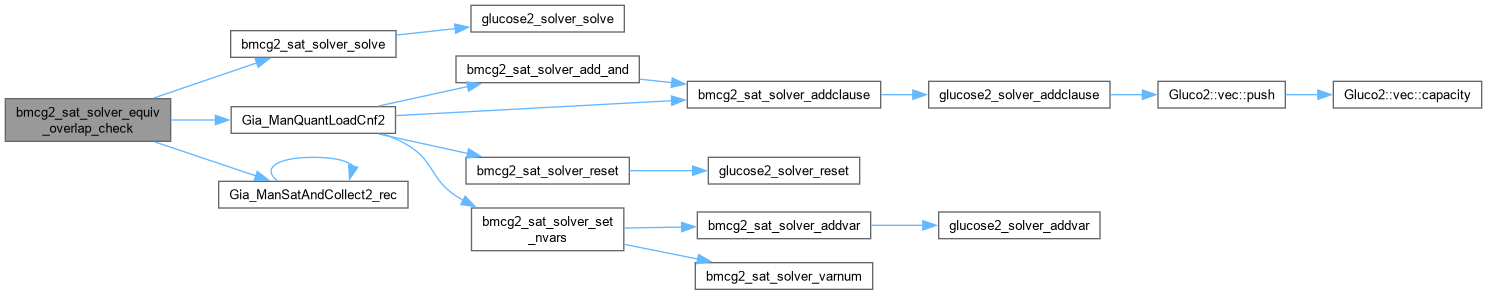
| int bmcg2_sat_solver_equiv_overlap_check | ( | bmcg2_sat_solver * | pSat, |
| Gia_Man_t * | p, | ||
| int | iLit0, | ||
| int | iLit1, | ||
| int | fEquiv ) |
Function*************************************************************
Synopsis [Checks equivalence or intersection of two nodes.]
Description []
SideEffects []
SeeAlso []
Definition at line 1432 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_final | ( | bmcg2_sat_solver * | s, |
| int ** | ppArray ) |
Definition at line 180 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_jftr | ( | bmcg2_sat_solver * | s | ) |
Definition at line 371 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_learntnum | ( | bmcg2_sat_solver * | s | ) |
Definition at line 263 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_mark_cone | ( | bmcg2_sat_solver * | s, |
| int | var ) |
| void bmcg2_sat_solver_markapprox | ( | bmcg2_sat_solver * | s, |
| int | v0, | ||
| int | v1, | ||
| int | nlim ) |
Definition at line 235 of file AbcGlucose2.cpp.


| int bmcg2_sat_solver_minimize_assumptions | ( | bmcg2_sat_solver * | s, |
| int * | plits, | ||
| int | nlits, | ||
| int | pivot ) |
Definition at line 272 of file AbcGlucose2.cpp.

| void bmcg2_sat_solver_prelocate | ( | bmcg2_sat_solver * | s, |
| int | var_num ) |
Definition at line 396 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_print_sop | ( | Gia_Man_t * | p | ) |
Definition at line 1041 of file AbcGlucose2.cpp.

| void bmcg2_sat_solver_print_sop_lit | ( | Gia_Man_t * | p, |
| int | Lit ) |
Definition at line 1047 of file AbcGlucose2.cpp.
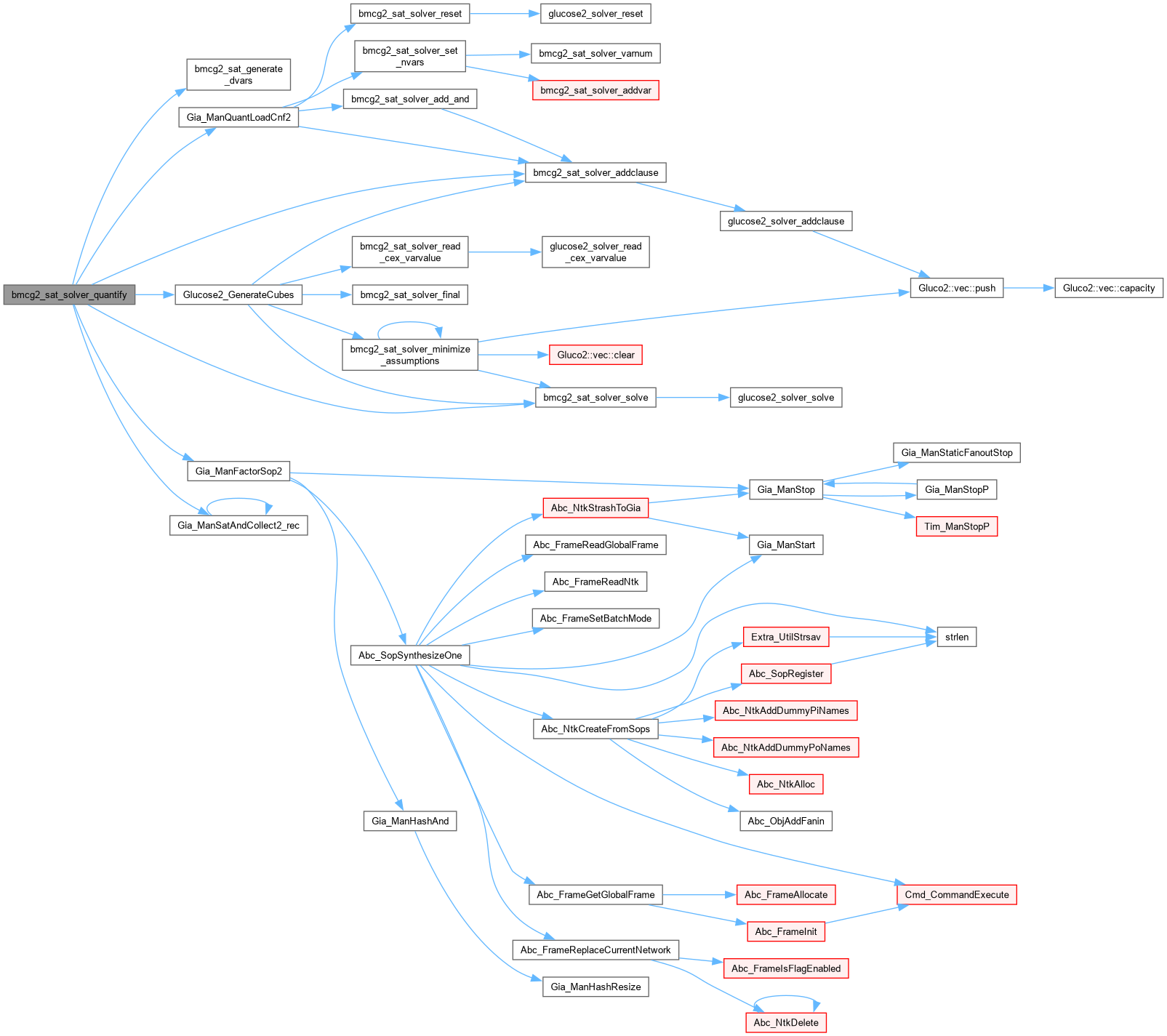
| int bmcg2_sat_solver_quantify | ( | bmcg2_sat_solver * | pSats[], |
| Gia_Man_t * | p, | ||
| int | iLit, | ||
| int | fHash, | ||
| int(* | pFuncCiToKeep )(void *, int), | ||
| void * | pData, | ||
| Vec_Int_t * | vDLits ) |
Definition at line 1274 of file AbcGlucose2.cpp.

| int bmcg2_sat_solver_quantify2 | ( | Gia_Man_t * | p, |
| int | iLit, | ||
| int | fHash, | ||
| int(* | pFuncCiToKeep )(void *, int), | ||
| void * | pData, | ||
| Vec_Int_t * | vDLits ) |
Function*************************************************************
Synopsis [Performs SAT-based quantification.]
Description []
SideEffects []
SeeAlso []
Definition at line 1114 of file AbcGlucose2.cpp.
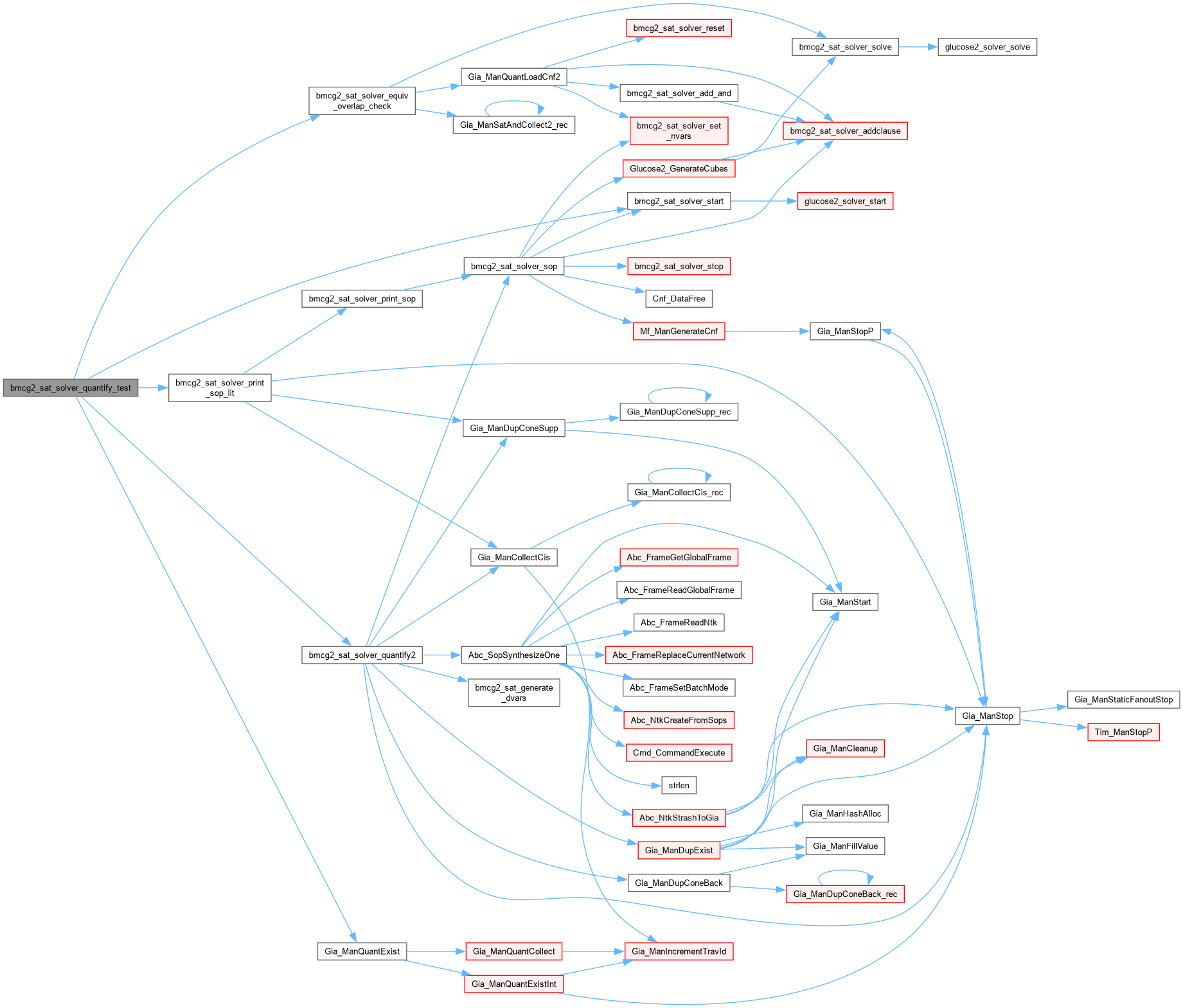
| int bmcg2_sat_solver_quantify_test | ( | bmcg2_sat_solver * | pSats[], |
| Gia_Man_t * | p, | ||
| int | iLit, | ||
| int | fHash, | ||
| int(* | pFuncCiToKeep )(void *, int), | ||
| void * | pData, | ||
| Vec_Int_t * | vDLits ) |
Definition at line 1402 of file AbcGlucose2.cpp.
| int * bmcg2_sat_solver_read_cex | ( | bmcg2_sat_solver * | s | ) |
Definition at line 221 of file AbcGlucose2.cpp.

| int bmcg2_sat_solver_read_cex_varvalue | ( | bmcg2_sat_solver * | s, |
| int | ivar ) |
Definition at line 225 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_reset | ( | bmcg2_sat_solver * | s | ) |
Definition at line 159 of file AbcGlucose2.cpp.

| void bmcg2_sat_solver_set_conflict_budget | ( | bmcg2_sat_solver * | s, |
| int | Limit ) |
Definition at line 247 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_set_jftr | ( | bmcg2_sat_solver * | s, |
| int | jftr ) |
Definition at line 376 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_set_nvars | ( | bmcg2_sat_solver * | s, |
| int | nvars ) |
Definition at line 191 of file AbcGlucose2.cpp.

| abctime bmcg2_sat_solver_set_runtime_limit | ( | bmcg2_sat_solver * | s, |
| abctime | Limit ) |
Definition at line 240 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_set_stop | ( | bmcg2_sat_solver * | s, |
| int * | pstop ) |
Definition at line 230 of file AbcGlucose2.cpp.

| void bmcg2_sat_solver_set_var_fanin_lit | ( | bmcg2_sat_solver * | s, |
| int | var, | ||
| int | lit0, | ||
| int | lit1 ) |
| void bmcg2_sat_solver_setcallback | ( | bmcg2_sat_solver * | s, |
| void * | pman, | ||
| int(* | pfunc )(void *, int, int *) ) |
Definition at line 170 of file AbcGlucose2.cpp.

| int bmcg2_sat_solver_solve | ( | bmcg2_sat_solver * | s, |
| int * | plits, | ||
| int | nlits ) |
Definition at line 175 of file AbcGlucose2.cpp.

Definition at line 998 of file AbcGlucose2.cpp.
| bmcg2_sat_solver * bmcg2_sat_solver_start | ( | ) |
MACRO DEFINITIONS ///.
Function*************************************************************
Synopsis [Wrapper APIs to calling from ABC.]
Description []
SideEffects []
SeeAlso []
Definition at line 151 of file AbcGlucose2.cpp.

| void bmcg2_sat_solver_start_new_round | ( | bmcg2_sat_solver * | s | ) |
Definition at line 386 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_stop | ( | bmcg2_sat_solver * | s | ) |
Definition at line 155 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_var_is_elim | ( | bmcg2_sat_solver * | s, |
| int | v ) |
Definition at line 204 of file AbcGlucose2.cpp.
| void bmcg2_sat_solver_var_set_frozen | ( | bmcg2_sat_solver * | s, |
| int | v, | ||
| int | freeze ) |
Definition at line 210 of file AbcGlucose2.cpp.
| int bmcg2_sat_solver_varnum | ( | bmcg2_sat_solver * | s | ) |
| int Gia_ManCiIsToKeep2 | ( | void * | pData, |
| int | i ) |
Definition at line 1371 of file AbcGlucose2.cpp.

Definition at line 1255 of file AbcGlucose2.cpp.

| void Gia_ManQuantLoadCnf2 | ( | Gia_Man_t * | p, |
| Vec_Int_t * | vObjsUsed, | ||
| bmcg2_sat_solver * | pSats[] ) |
Definition at line 1225 of file AbcGlucose2.cpp.
| int Gia_ManSatAndCollect2_rec | ( | Gia_Man_t * | p, |
| int | iObj, | ||
| Vec_Int_t * | vObjsUsed, | ||
| Vec_Int_t * | vCiVars ) |
Function*************************************************************
Synopsis [Performs SAT-based quantification.]
Description []
SideEffects []
SeeAlso []
Definition at line 1206 of file AbcGlucose2.cpp.


| void Glucose2_CheckTwoNodesTest | ( | Gia_Man_t * | p | ) |
Definition at line 1477 of file AbcGlucose2.cpp.
| Vec_Str_t * Glucose2_GenerateCubes | ( | bmcg2_sat_solver * | pSat[2], |
| Vec_Int_t * | vCiSatVars, | ||
| Vec_Int_t * | vVar2Index, | ||
| int | CubeLimit ) |
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 940 of file AbcGlucose2.cpp.

| void glucose2_markapprox | ( | Gluco2::SimpSolver * | S, |
| int | v0, | ||
| int | v1, | ||
| int | nlim ) |
Definition at line 135 of file AbcGlucose2.cpp.

| void glucose2_print_stats | ( | SimpSolver & | s, |
| abctime | clk ) |
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 766 of file AbcGlucose2.cpp.
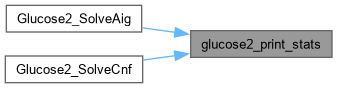
| void Glucose2_QuantifyAigTest | ( | Gia_Man_t * | p | ) |
Definition at line 1375 of file AbcGlucose2.cpp.
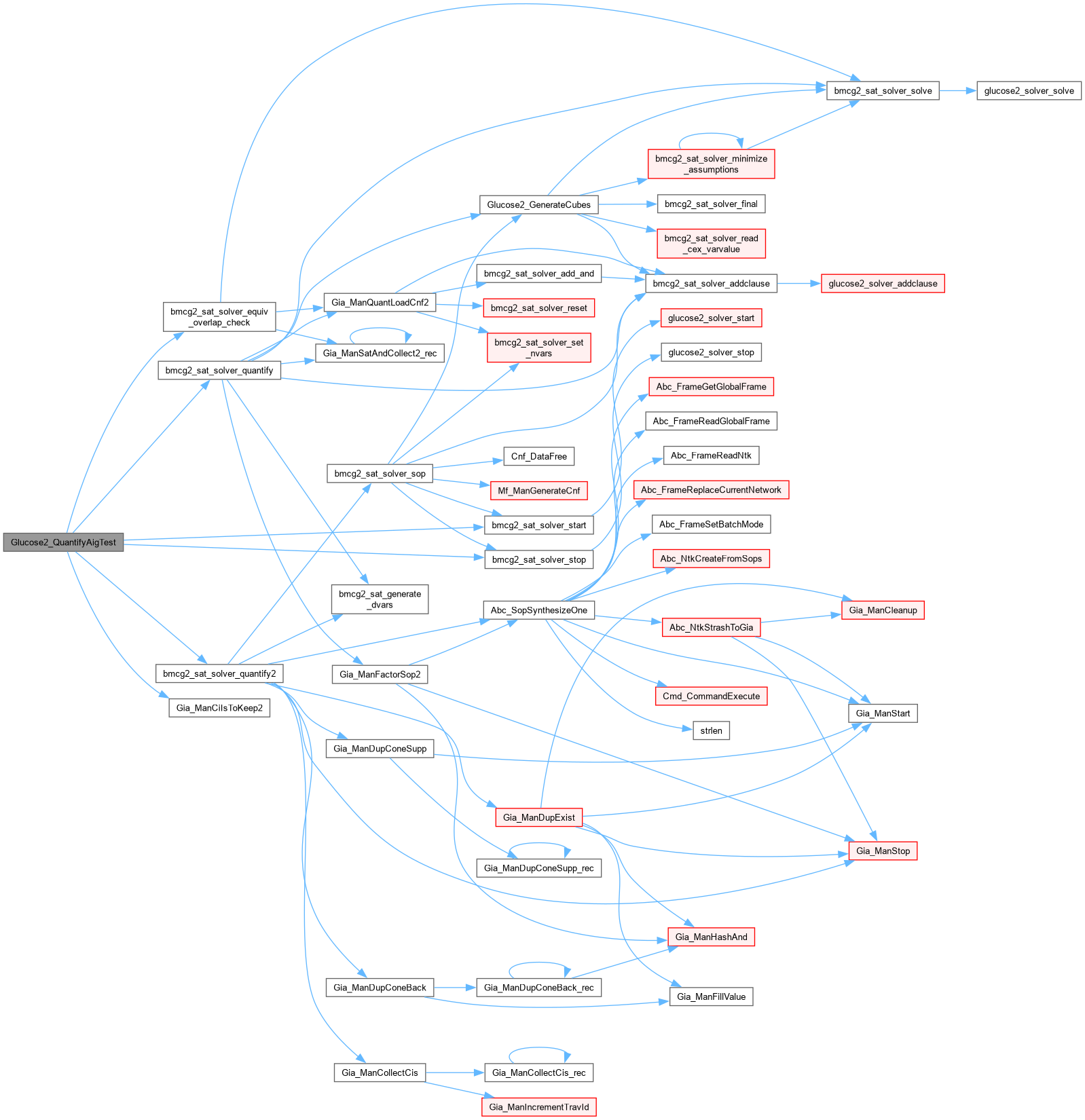
| int Glucose2_SolveAig | ( | Gia_Man_t * | p, |
| Glucose2_Pars * | pPars ) |
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 1505 of file AbcGlucose2.cpp.
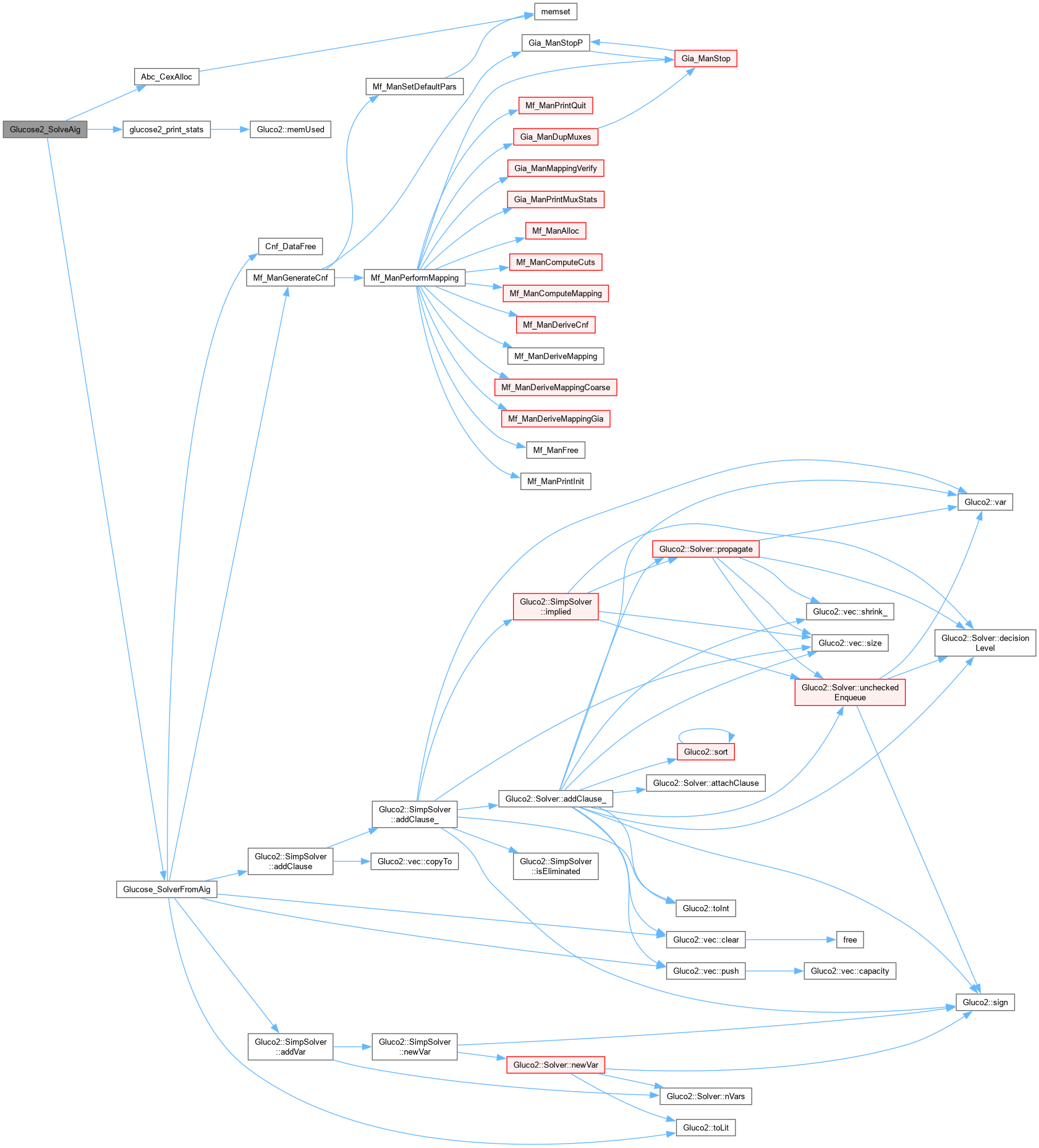
| void Glucose2_SolveCnf | ( | char * | pFileName, |
| Glucose2_Pars * | pPars ) |
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 848 of file AbcGlucose2.cpp.
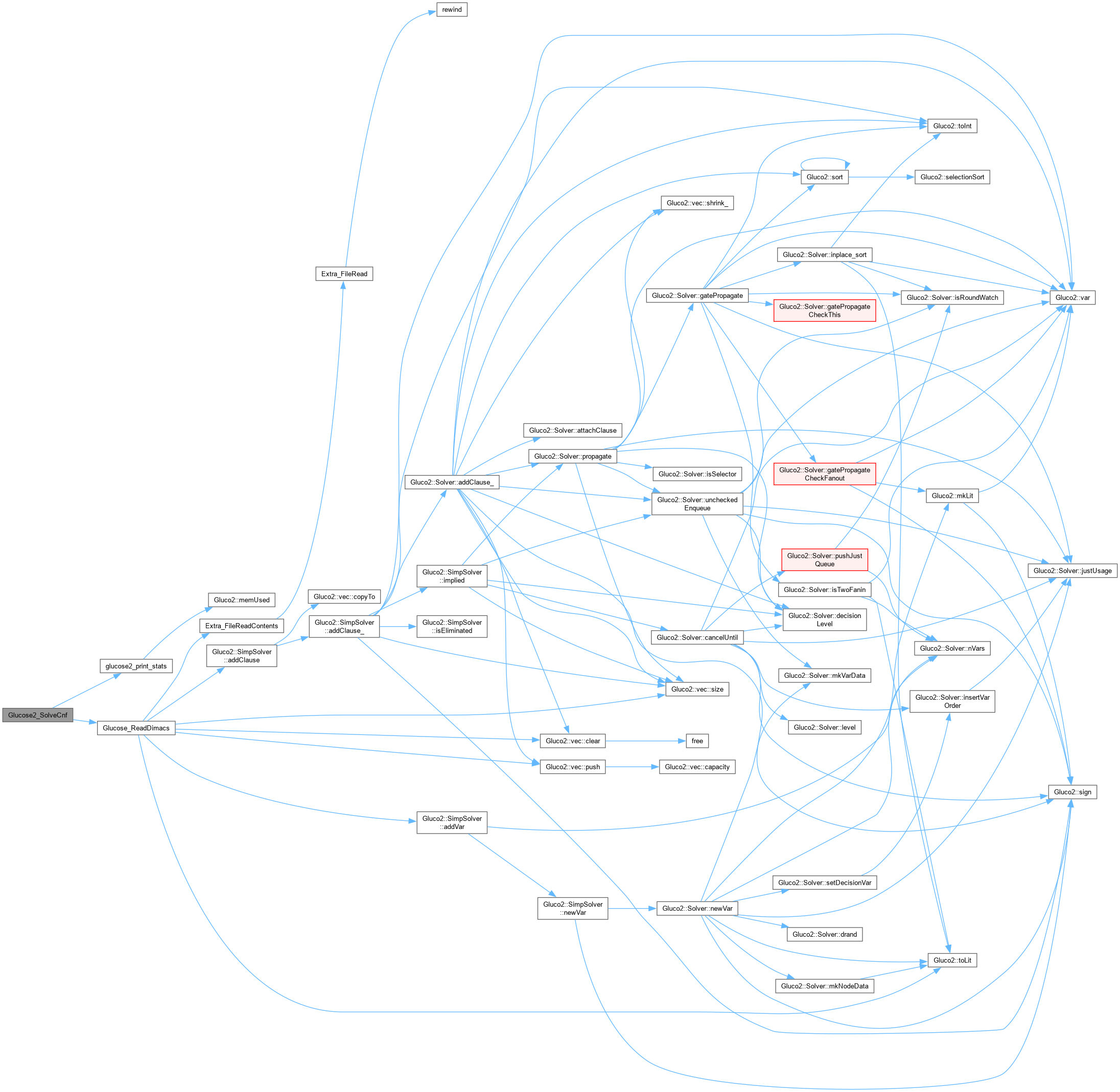
| int glucose2_solver_addclause | ( | Gluco2::SimpSolver * | S, |
| int * | plits, | ||
| int | nlits ) |
Definition at line 78 of file AbcGlucose2.cpp.

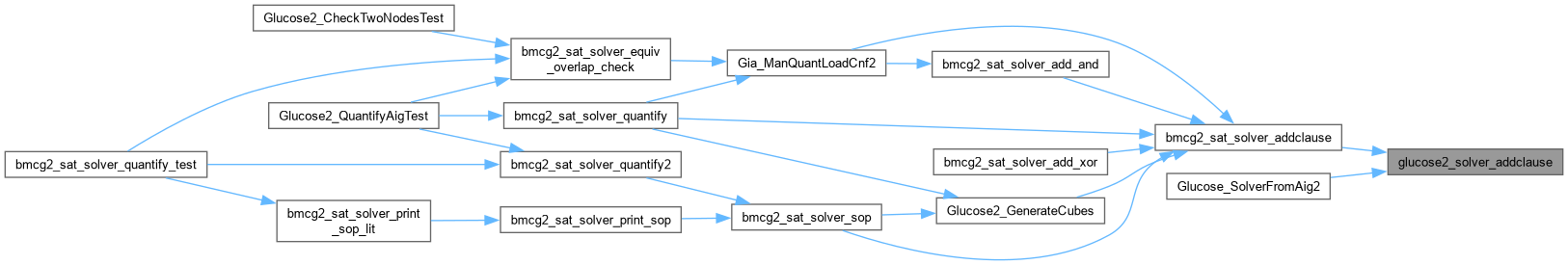
| int glucose2_solver_addvar | ( | Gluco2::SimpSolver * | S | ) |
Definition at line 114 of file AbcGlucose2.cpp.
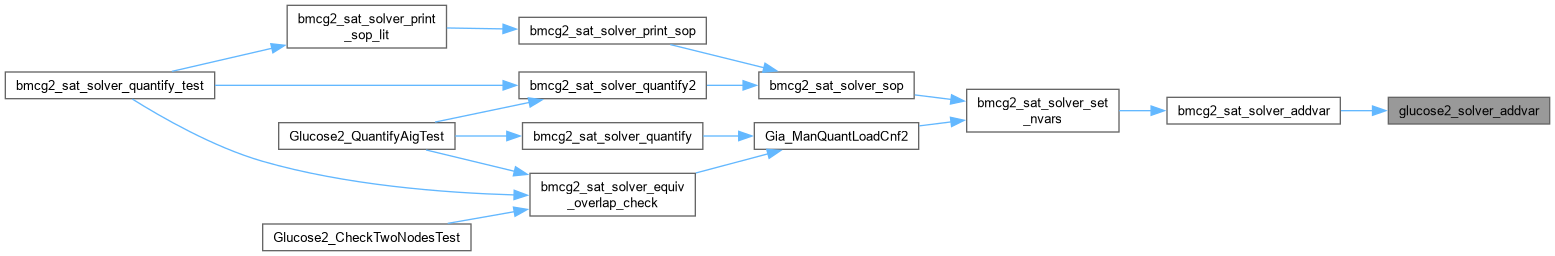
| int * glucose2_solver_read_cex | ( | Gluco2::SimpSolver * | S | ) |
Definition at line 120 of file AbcGlucose2.cpp.

| int glucose2_solver_read_cex_varvalue | ( | Gluco2::SimpSolver * | S, |
| int | ivar ) |
| void glucose2_solver_reset | ( | Gluco2::SimpSolver * | S | ) |
Definition at line 73 of file AbcGlucose2.cpp.
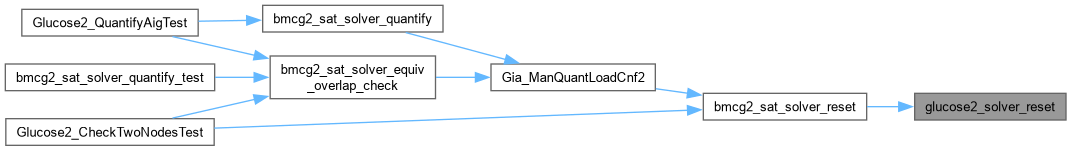
| void glucose2_solver_setcallback | ( | Gluco2::SimpSolver * | S, |
| void * | pman, | ||
| int(* | pfunc )(void *, int, int *) ) |
Definition at line 93 of file AbcGlucose2.cpp.

| void glucose2_solver_setstop | ( | Gluco2::SimpSolver * | S, |
| int * | pstop ) |
Definition at line 130 of file AbcGlucose2.cpp.

| int glucose2_solver_solve | ( | Gluco2::SimpSolver * | S, |
| int * | plits, | ||
| int | nlits ) |
Definition at line 100 of file AbcGlucose2.cpp.
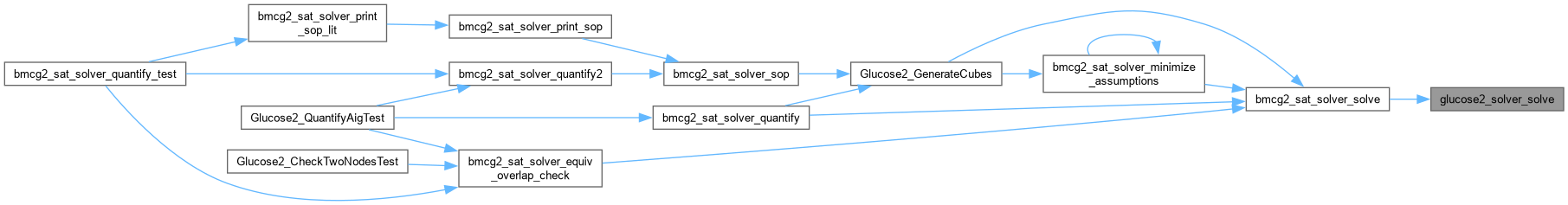
| SimpSolver * glucose2_solver_start | ( | ) |
FUNCTION DEFINITIONS ///.
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 61 of file AbcGlucose2.cpp.

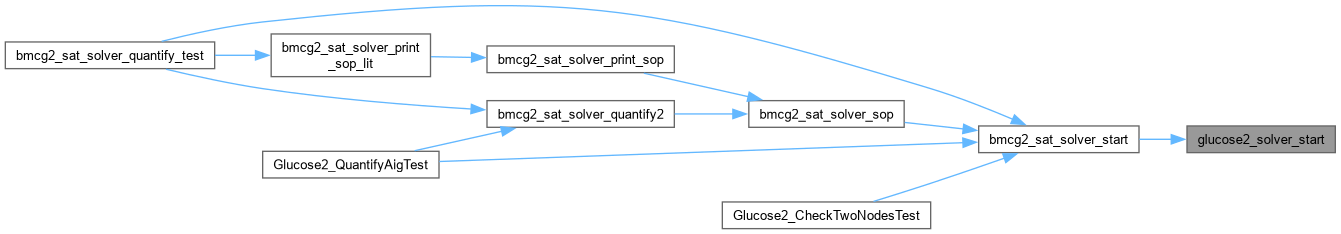
| void glucose2_solver_stop | ( | Gluco2::SimpSolver * | S | ) |
Definition at line 68 of file AbcGlucose2.cpp.
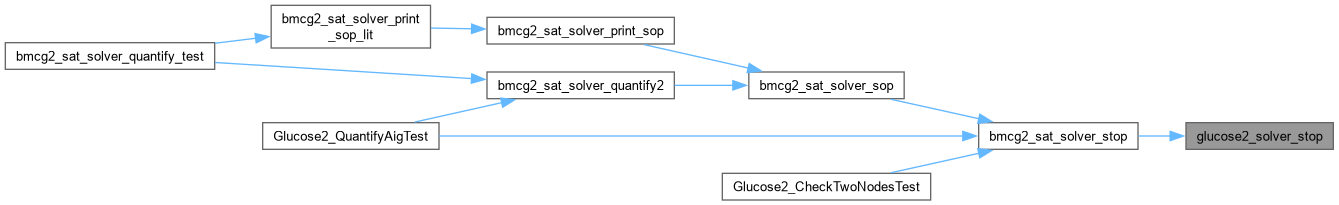
| void Glucose_ReadDimacs | ( | char * | pFileName, |
| SimpSolver & | s ) |
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 798 of file AbcGlucose2.cpp.
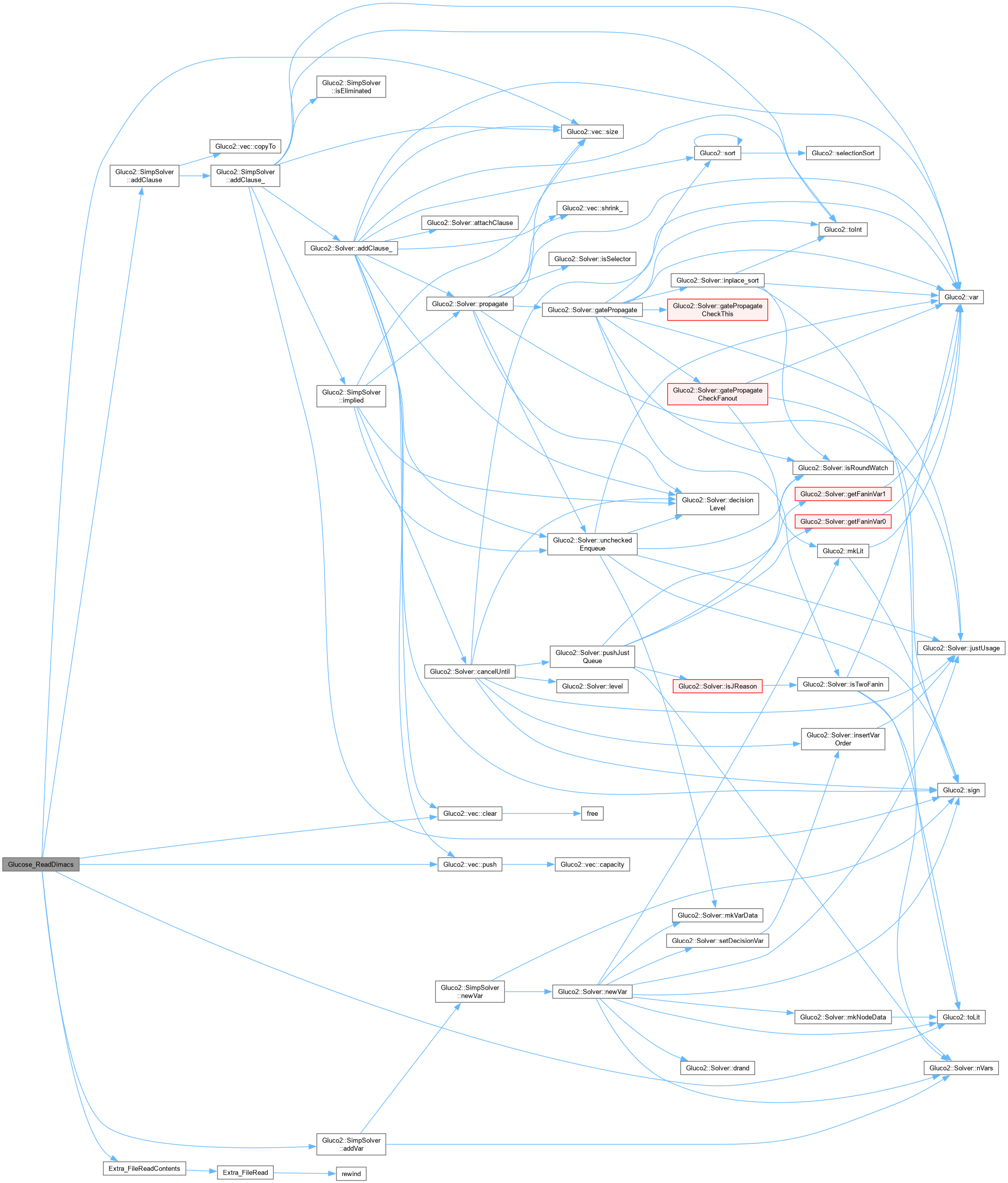

| Vec_Int_t * Glucose_SolverFromAig | ( | Gia_Man_t * | p, |
| SimpSolver & | s ) |
Function*************************************************************
Synopsis []
Description []
SideEffects []
SeeAlso []
Definition at line 894 of file AbcGlucose2.cpp.
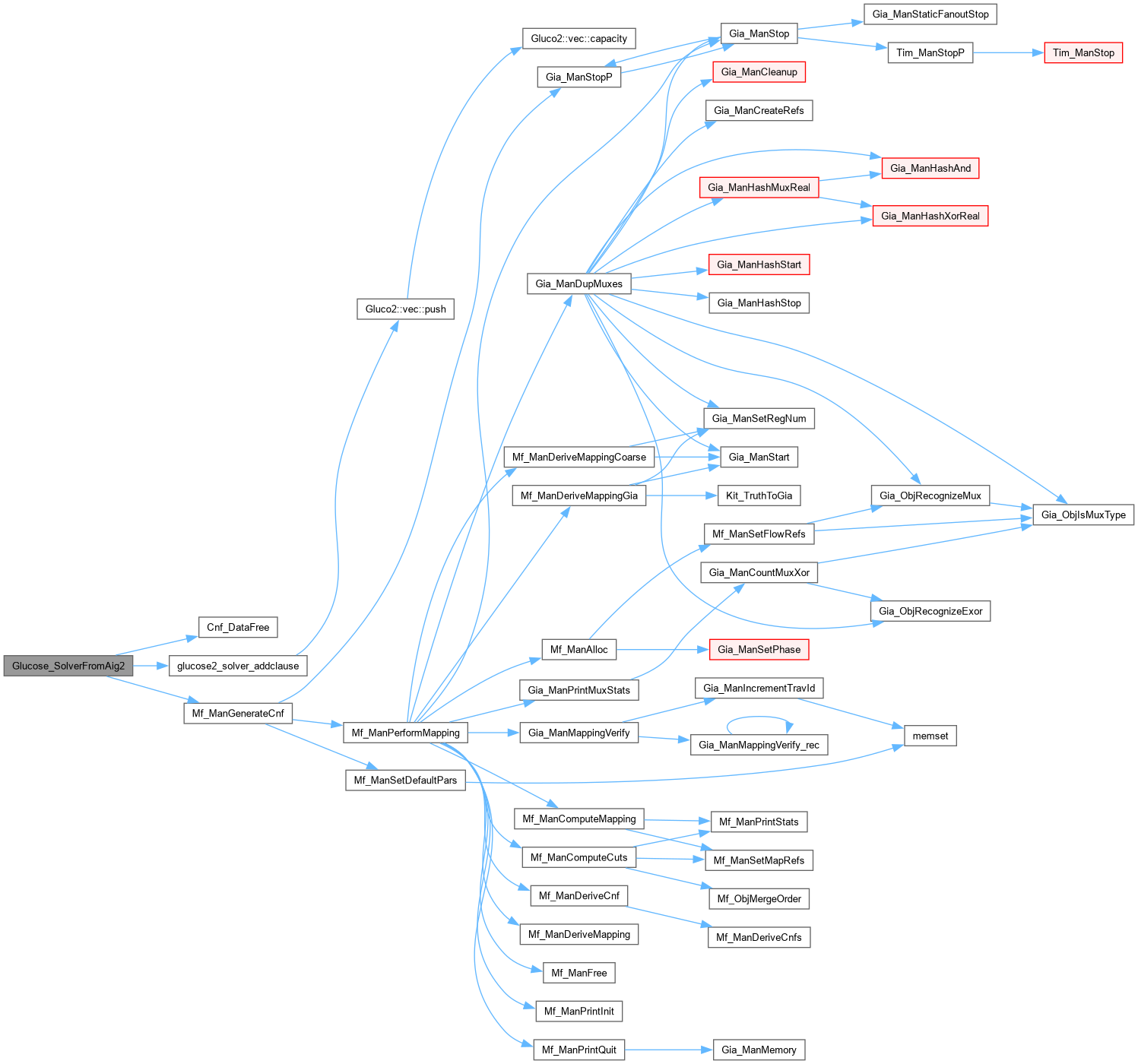
| Vec_Int_t * Glucose_SolverFromAig2 | ( | Gia_Man_t * | p, |
| SimpSolver & | S ) |
Definition at line 914 of file AbcGlucose2.cpp.